

The graphs shown below ilustrate how AFAL uncovers the most frequent (y-axis, percentages) amino acids found in the coordination sphere of a certain ligand, as defined by the user, when applying the following filters:
Analysis of aminoacid surrounding the ATP molecule


Jmol box: we show some example of interaction between the protein phosphoenolpyruvate carboxykinase (pepck) with the ATP (PDB code 1AQ2) to illustrate the analysis performed by AFAL.
As shown in the chart, Gly, Lys, Asp, Ser, Arg, Glu and Thr are one the most frequent amino acids identified by the AFAL algorithm that surround ATP in the 564 available ATP-binding proteins from PDB and in different source organisms. These are typically present in the P-loop motif [GXXXXGKT(S)], responsible the recognition of phosphate group. (Ramakrishnan, et al., 2002. Protein Engineering, 15(10), 783–798).

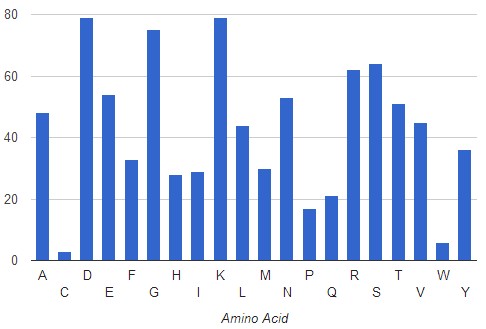
Narrowing down the search to specific protein families, additional tendencies in the use of certain residues for the coordination of ATP emerge. AFAL correctly identifies the three conserved residues (Gly, Lys, Asp) that define the P-loop motif described for the Transferase family and, interestingly, also an increased presence of the positively charged residue Arginine potentially involved in adenine-protein cation-π interactions.

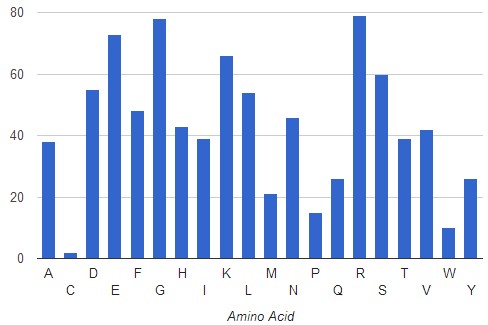
The analysis for the protein belonging to the Ligase family shows a similar tendency, although increased frequencies of Arg over P-loop motif residues suggest adenine-protein cation-π interactions are predominant in this protein family. Aromatic residues that define the A-loop are seldom found in the whole dataset (approximately 3%). These findings are in agreement with previous studies and validate the utility of AFAL for analysis of protein-ligand interaction patterns. It also demonstrates the power of AFAL to find novel amino acid – ligand patterns .