Presentation
GSP4PDB is a bioinformatics web tool that lets the users visualize, search and explore protein-ligand structural patterns inside the Protein Data Bank. (PDB).
The novel feature of GSP4PDB is that a protein-ligand structural pattern is graphically represented as a graph such that the nodes represent protein’s components (amino acids and ligands) and the edges represent structural relationships (e.g. distance relationships). Such abstract representation is called a Graph-based Structural Pattern (GSP).
Once the user has "drawn" the GSP, it is transformed into an SQL query, and searched in a PostgreSQL database containing PDB data. The results of the search are shown in textual or graphical form, depending of the version of of GSP4PDB.
The first version of GSP4PDB is described in:(A graph-based approach for querying protein-ligand structural patterns. R. Angles and M. Arenas. 6th International Work-Conference on Bioinformatics and Biomedical Engineering (IWBBIO), LNBI 10813. Granada, Spain, April 2018) .
Protein-ligand structural pattern
The notion of structural pattern is used to describe a three-dimensional "structure" or "shape" that occurs in the secondary structure of a protein.
We define a protein-ligand structural pattern as the combination of a ligand and a group of amino acids, whose three-dimensional distribution could be determined by three types of relationships:
Distance between two amino acids.
Distance between an amino and the ligand.
Order of precedence (in the sequence) of an amino respect to other amino acid.
For instance, a zinc finger is a protein-ligand structural pattern where a zinc atom (the ligand) is surrounded by cysteine and histidine residues (the amino acids).
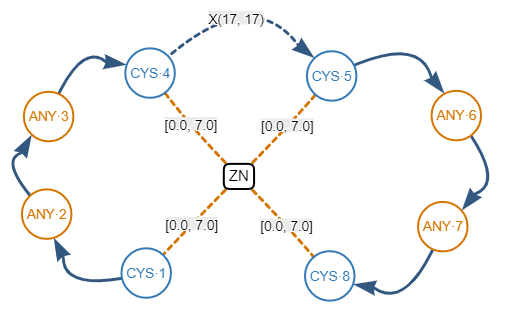
C-x(2)-C-x(17)-C-x(2)-C
Graph-based structural pattern (GSP) is a labeled property graph where nodes and edges can contain key-values pairs representing their propeties (or attributes). For this case (GSP4PDB) four types of nodes are allowed:
Amino-acid-nodes
Any-amino-acid-nodes
Ligand-nodes
Any-ligand-nodes
Additionaly, nodes can be connected by three types of edges:
Distance-edges
Next-edges
Gap-edges
Protein data storage
GSP4PDB uses a PostgreSQL database system (v 9.4) for storing and managing protein data obtained from the PDB repository. The database is formed by the following relational tables: protein, chain, standard_amino, aminoacid, ligand, atom_amino, atom_ligand, distance_amino_amino, distance_ligand_amino, next_amino_amino, protein_cath.
The core of the database is given by the tables protein, chain, standard_amino, aminoacid, ligand, atom_amino and atom_ligand. The Table standard_amino contains information about 20 standard amino acids, plus an "undefined" amino acid, whose symbol and abreviation are "U" and "UND" respectively. The table protein_cath contains information about the CATH classification.
It is possible to see that a large part of database corresponds to the information related to distances, in particular, distances between each pair of amino acids. Note that the number of chains is not equal to the number of proteins, as some PDB files contain nucleic acid data. In such case we maintain basic information about the nucleic acid in the "Protein" table.
In practice, just the relational tables protein, protein_cath, standard_amino, distance_amino_amino, distance_ligand_amino and next_amino_amino are used to search graph-based structural patterns. The rest have been included to maintain additional information, and future developments.
Fore more information about the tables and statistics, please visit SQL4PDB.
Query processing
The query procesing consists in transforming a graph-based structural pattern (GSP) in queries. In general terms, the method generates a SQL query expression for each node-edge-node structure in the graph pattern. The final SQL query, expressing the complete graph pattern, is the compositions of all the sub-expressions.
Fore more information about the query processing, please visit Patterns to SQL queries transformation..
Versions
About...
GSP4PDB is part of the services provided by the Bioinformatic Group of the University of Talca
Send your questions, suggestions or comments to the email contact: rangles@gmail.com
© 2018 Designed by Diego Cisterna , Roberto García and Ph.D. Renzo Angles
Send your questions, suggestions or comments to the email contact: rangles@gmail.com
© 2018 Designed by Diego Cisterna , Roberto García and Ph.D. Renzo Angles