Presentation
Well defined biomacromolecular patterns such as binding sites between a region of a protein and a ligand regulate certain factors that make possible the origin of many important biological phenomena. GSP4PDB is a user-friendly web application that lets the users design, search and analyze structural patterns inside biomacromolecules in a graphic way. This application uses the Protein Data Bank (PDB) to obtain detailed structural information.
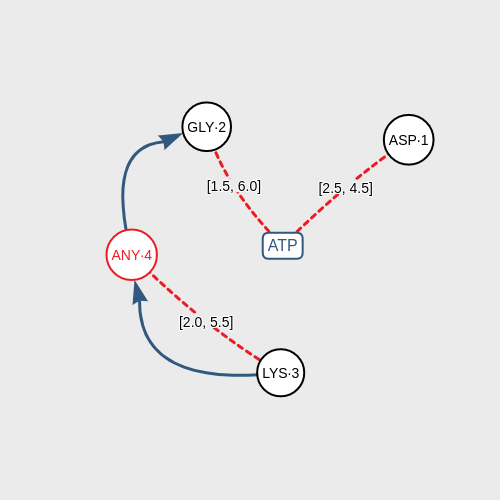
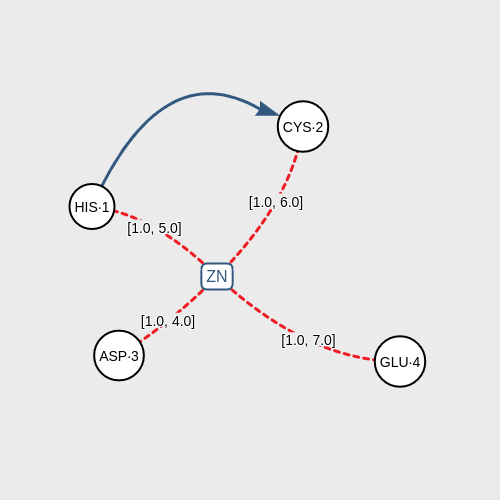
The program receives a graph-based pattern formed by graphic components that represent a ligand and the amino acids around it. The pattern also includes some associations that condition the structure: distances between the ligand and the amino acids, distances between amino acids, and sequence of amino acids in the chain.
The user can determine what type of ligand want to analyze and include fixed amino acids but also can include amino acids 'any'. They can take the value of any amino acid, although the user can restrict them to a specific classification according to their polarity.
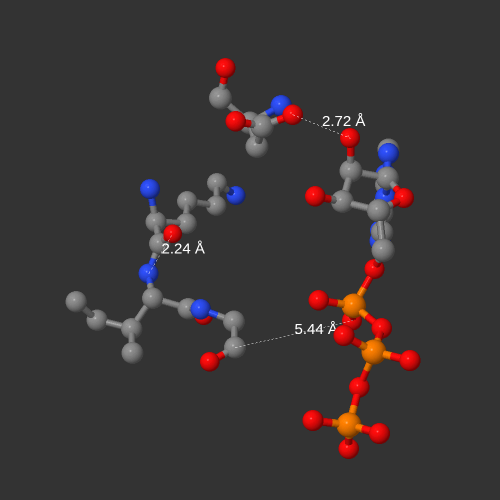
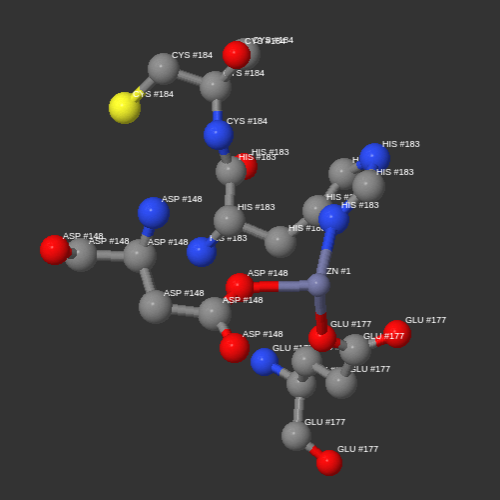
Finally, the program looks for coincidences inside each macromolecule in the PDB. The results of the search are shown in textual form and graphically by a 3D viewer. This analysis allows comparing proteins, DNA or RNA where the structure exists and evaluating how the amino acids surround the ligand in each coincidence.
Instructions
In the work panel, click on the LIGAND button to add a ligand of interest. This is mandatory.
Add amino acids by clicking on the buttons AMINO ACID and ANY AMINO as you like. You must add at least one of these obligatorily.
Connect all the components that you added to the panel using the buttons DISTANCE and NEXT.
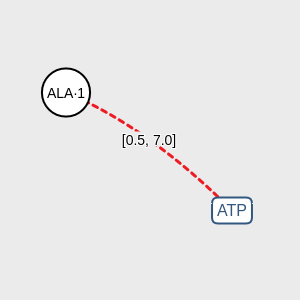
Click on DISTANCE to associate two components according to their proximity measured in angstroms (Å). Default range of distance is [0.5, 7.0]. You can modify this range doing double-click on the red lines. You have to use this association to connect the ligand and one amino acid obligatorily.
Click on NEXT to associate two amino acids according to the sequence of the chain of amino acids inside of the biomacromolecule.After you have designed the structural pattern correctly, click on the SEARCH button and the application will start looking for coincidences of your pattern in PDB.
Observe the results of the search shown individually on the panel of the right. You can analyze the structure of each coincidence in the 3D viewer by clicking the 3D STRUCTURE buttons.
The Tool
Make a structural pattern of amino acids and a ligand
Search results
Examples
About...
When using GSP4PDB, please consult (and cite) the following reference:
Diego Cisterna. Diseño de una interfaz gráfica para búsqueda de patrones estructurales en el Protein Data Bank. Universidad de Talca, Julio 2017.
Send your questions, suggestions or comments to the email contact: rangles@utalca.cl
© 2017 Designed by Diego Cisterna and Ph.D. Renzo Angles